Extraction of Robust Voids and Pockets in Proteins
The Eurographics Conference on Visualization, 2013
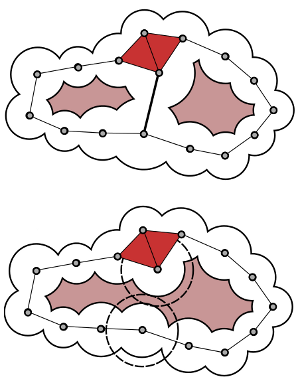
Abstract:
Topological methods have been successfully used to identify features in scalar fields and to measure their importance. Voids and pockets in a protein refer to empty spaces that are enclosed by the protein molecule. Existing methods to compute, measure, and visualize the voids and pockets in a protein molecule are sensitive to inaccuracies in the empirically determined atomic radii. This paper presents a topological framework that enables robust computation and visualization of these structures. Given a fixed set of atoms, voids and pockets are represented as subsets of the weighted Delaunay triangulation of atom centers. A novel notion of (ϵ,π)-stable voids helps identify voids that are stable even after perturbing the atom radii by a small value. An efficient method is described to compute these stable voids for a given input pair of values (ϵ,π).
Download Paper DOI
To cite: Raghavendra Sridharamurthy, Harish Doraiswamy, Siddharth Patel, Raghavan Varadarajan and Vijay Natarajan "Extraction of Robust Voids and Pockets in Proteins" EuroVis (Short Papers), 2013 10.2312/PE.EUROVISSHORT.EUROVISSHORT2013.067-071
BibTeX:
@inproceedings{SridharamurthyetalEuroVis2013, url = {http://diglib.eg.org/handle/10.2312/PE.EuroVisShort.EuroVisShort2013.067-071}, author = {Sridharamurthy, Raghavendra and Doraiswamy, Harish and Patel, Siddharth and Varadarajan, Raghavan and Natarajan, Vijay}, keywords = {I.3.6 [Computer Graphics], Computer Graphics, Methodology and Techniques}, title = {{Extraction of Robust Voids and Pockets in Proteins}}, conference = {EuroVis - Short Papers}, publisher = {The Eurographics Association}, year = {2013}} 