Identification and Quantification of Important Voids and Pockets in Proteins
Indian Institute of Science, 2013
Abstract:
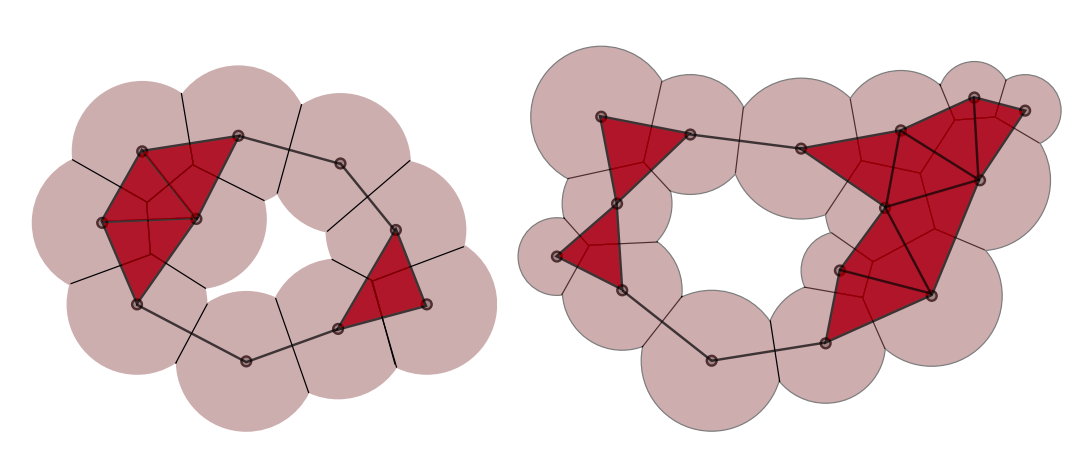
Many methods of analyzing both the physical and chemical behavior of proteins require information about its structure and stability. Also various other parameters such as energy function, solvation, hydrophobic/hydrophilic effects, surface area and volumes too play an important part in such analysis. The contribution of cavities to these parameters are very important. Existing methods to compute and measure cavities are limited by the inherent inaccuracies in the method of acquisition of data through x-ray crystallography and uncertainities in computation of radii of atoms. We present a topological framework that enables robust computation and visualization of these structures. Given a fixed set of atoms, voids and pockets are represented as subsets of the weighted Delaunay triangulation of atom centers. A novel notion of (ϵ,π)-stable voids helps identify voids that are stable even after perturbing the atom radii by a small value. An efficient method is described to compute these stable voids for a given input pair of values (ϵ,π). We also provide an implementation to visualize, explore (ϵ,π)-stable voids and also calculate various properties such as volumes, surface areas of the proteins and also of the cavities.
Download Paper DOI
To cite: Raghavendra G S "Identification and Quantification of Important Voids and Pockets in Proteins", M.Sc(Engg) Thesis, Indian Institute of Science, 2013
BibTeX:
@mastersthesis{SridhaamurthyMScEngg2013,
title={{Identification and Quantification of Important Voids and Pockets in Proteins}},
author={Raghavendra G S},
year={2013},
school={Indian Institute of Science},
howpublished = {\url{https://etd.iisc.ac.in/handle/2005/3345}},
address={Bengaluru}
} 